Scroll to:
GATCGGenerator: New Software for Generation of Quasirandom Nucleotide Sequences
https://doi.org/10.23947/2687-1653-2023-23-3-296-306
Abstract
Introduction. In recent decades, knowledge about DNA has been increasingly used to solve biological problems (calculations using DNA, long-term storage of information). Principally, we are talking about cases when it is required to select artificial nucleotide sequences. Special programs are used to create them. However, existing generators do not take into account the physicochemical properties of DNA and do not allow obtaining sequences with a pronounced “non-biological” structure. In fact, they generate sequences by distributing nucleotides randomly. The objective of this work is to create a generator of quasirandom sequences with a special nucleotide structure. It should take into account some physicochemical features of nucleotide structures, and it will be involved in storing non-biological information in DNA.
Materials and Methods. A new GATCGGenerator software for generating quasirandom sequences of nucleotides was described. It was presented as SaaS (from “software as a service”), which provided its availability from various devices and platforms. The program generated sequences of a certain structure taking into account the guanine-cytosine (GC) composition and the content of dinucleotides. The performance of the new program algorithm was presented. The requirements for the generated nucleotide sequences were set using a chat in Telegram, the interaction with the user was clearly shown. The differences between the input parameters and the specific nucleotide structures obtained as a result of the program were determined and generalized. Also, the time costs of generating sequences for different input data were given in comparison. Short sequences differing in type, length, GC composition and dinucleotide content were studied. The tabular form shows how the input and output parameters are correlated in this case.
Results. The developed software was compared to existing nucleotide sequence generators. It has been established that the generated sequences differ in structure from the known DNA sequences of living organisms, which means that they can be used as auxiliary or masking oligonucleotides suitable for molecular biological manipulations (e.g., amplification reactions), as well as for storing non-biological information (images, texts, etc.) in DNA molecules. The proposed solution makes it possible to form specific sequences from 20 to 5 000 nucleotides long with a given number of dinucleotides and without homopolymer fragments. More stringent generation conditions remove known limitations and provide the creation of quasirandom sequences of nucleotides according to specified input parameters. In addition to the number and length of sequences, it is possible to determine the GC composition, the content of dinucleotides, and the nature of the nucleic acid (DNA or RNA) in advance. Examples of short sequences differing in length, GC composition and dinucleotide content are given. The obtained 30-nucleotide sequences were tested. The absence of 100 % homology with known DNA sequences of living organisms was established. The maximum coincidence was observed for the generated sequences with a length of 25 nucleotides (similarity of about 80 %). Thus, it has been proved that GATCGGenerator can generate non-biological nucleotide sequences with high efficiency.
Discussion and Conclusion. The new generator provides the creation of nucleotide sequences in silico with a given GC composition. The solution makes it possible to exclude homopolymer fragments, which improves qualitatively the physicochemical stability of sequences.
Keywords
For citations:
Kiryanova O.Yu., Garafutdinov R.R., Gubaydullin I.M., Chemeris A.V. GATCGGenerator: New Software for Generation of Quasirandom Nucleotide Sequences. Advanced Engineering Research (Rostov-on-Don). 2023;23(3):296-306. https://doi.org/10.23947/2687-1653-2023-23-3-296-306
Introduction. DNA is a unique biopolymer that provides storage, transmission and reproduction of genetic information in living organisms. DNA molecules consist of four types of nucleotides containing nitrogenous bases: adenine (A), guanine (G), cytosine (C), thymine (T). Their possible combinations provide nucleotide sequences forming functional genetic elements. In molecular biology and genetics, the basic investigations are carried out on nucleotide sequences of living organisms, but there is an increasing need to create artificial sequences, especially, when solving non-biological tasks (e.g., DNA calculations [1][2], storage in DNA [3], cryptography [4], DNA tags in hydrology [5], etc.).
It is expected that by the end of 2040, the volume of information will reach several yottabytes (1024), which requires its structuring and storage. Both of these processes affect significantly the consumption of energy resources, as well as the production of storage devices and peripheral devices (hard drives, solid-state drives). To store such an amount of information, more than 109 kg of extra pure silicon is required [6], which may not be enough. The solution is seen in using the principles of DNA to work with large-scale amounts of data.
Nucleotide sequences are easily digitized by assigning the corresponding binary codes to individual nucleotides
[7–11] or blocks of nucleotides [12–14]; therefore, text, graphic or multimedia files can be converted into nucleotide sequences [15–18]. Artificial nucleotide sequences can be made manually or generated using special software (DNA generators), depending on the tasks being solved. Some DNA generators were developed as independent applications, others — as part of software packages designed to solve general [19] 1, 2, 3, 4, 5 or specific tasks [20]. As a rule, DNA generators are developed on the basis of combinatorial approaches and produce random sequences of a given length of guanine-cytosine (GC) composition. However, such software solutions do not take into account the chemical properties of nucleotides and do not provide obtaining sequences with a certain structure (e.g., without homopolymer sites or long repeating motifs). Therefore, the sequences created by such generators cannot always be reproduced in the laboratory. Moreover, such sequences may be identical to DNA fragments existing in nature, which introduces ambiguity when trying to encode information of a non-biological nature.
The presented work is aimed at creating a generator of nucleotide sequences of a special structure that can be used when encoding text, graphic and other information in DNA molecules.
Materials and Methods. The criteria that should be kept in mind when creating sequences were defined. The need to vary the GC composition, set a certain number of dinucleotides, and exclude homopolymer sites in sequences was taken into account.
A team of authors has developed the GATCGGenerator program in Python 3.6 (Anaconda distribution)6. To create a bot7 in Telegram, Numpy 1.19 [21] and the Python GATCGGenerator library were used. The solution was provided as SaaS (from “software as a service”), which opened up the possibility of access from different devices and platforms.
Input parameters included the number of sequences, their length, GC composition, and dinucleotide content. The generator excluded repeats with a length of two nucleotides more than four times. The result was presented as a CSV file, which contained the following information: sequence, GC composition, and the number of all nucleotides.
Repeats and homopolymer fragments were stored as a separate list. First, a sequence of four elements was randomly generated (random.choice(nuk), where nuc = 'ACGT'). Then the search for repetitions was performed. If there was at least one item from the list, a new random generation was performed. Next, the GC and NN composition was calculated. If the NN composition did not match the user-defined range, the paired nucleotide was replaced randomly and the GC composition was recalculated. If the sequence matched the input parameters, it was written to a set of sequences.
Below is the operation of the program algorithm.
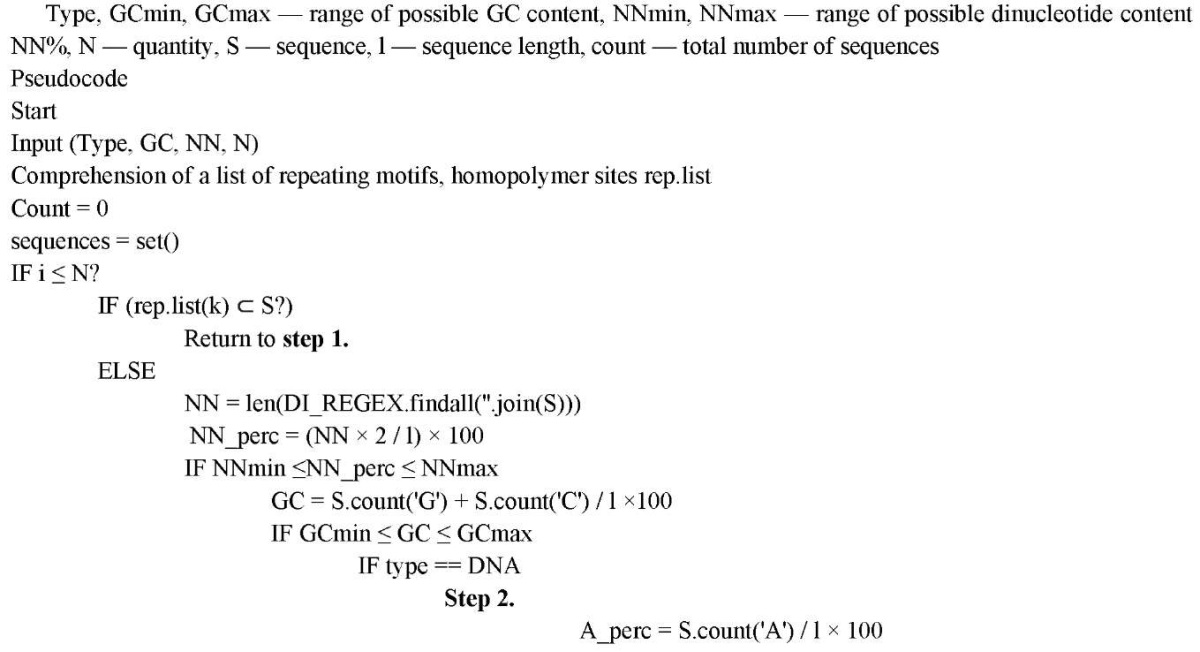
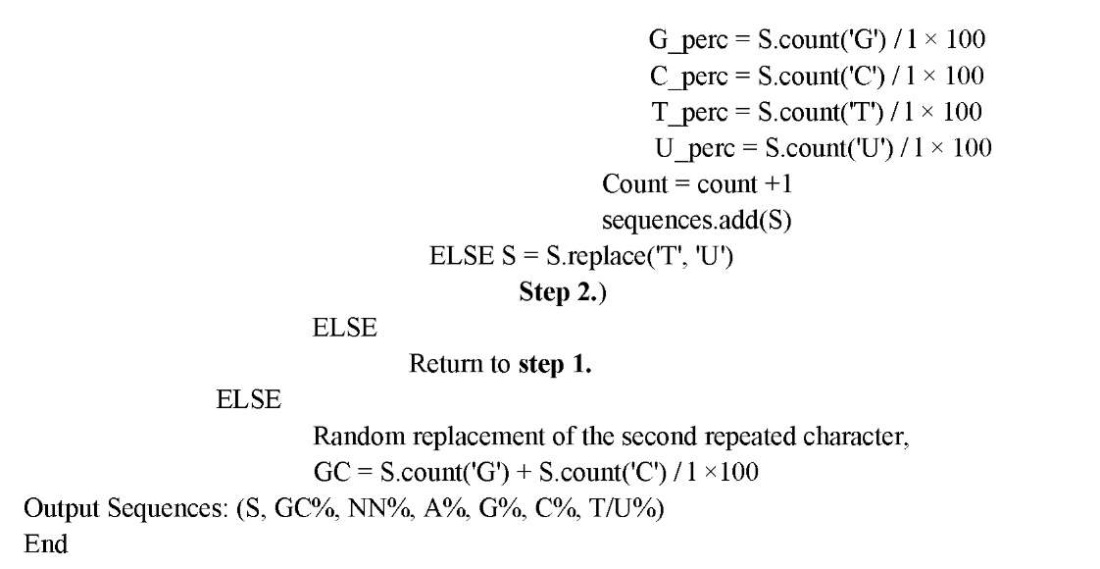
The requirements for the generated nucleotide sequences were set using Telegram chat. An example of user interaction is shown in Figure 1.
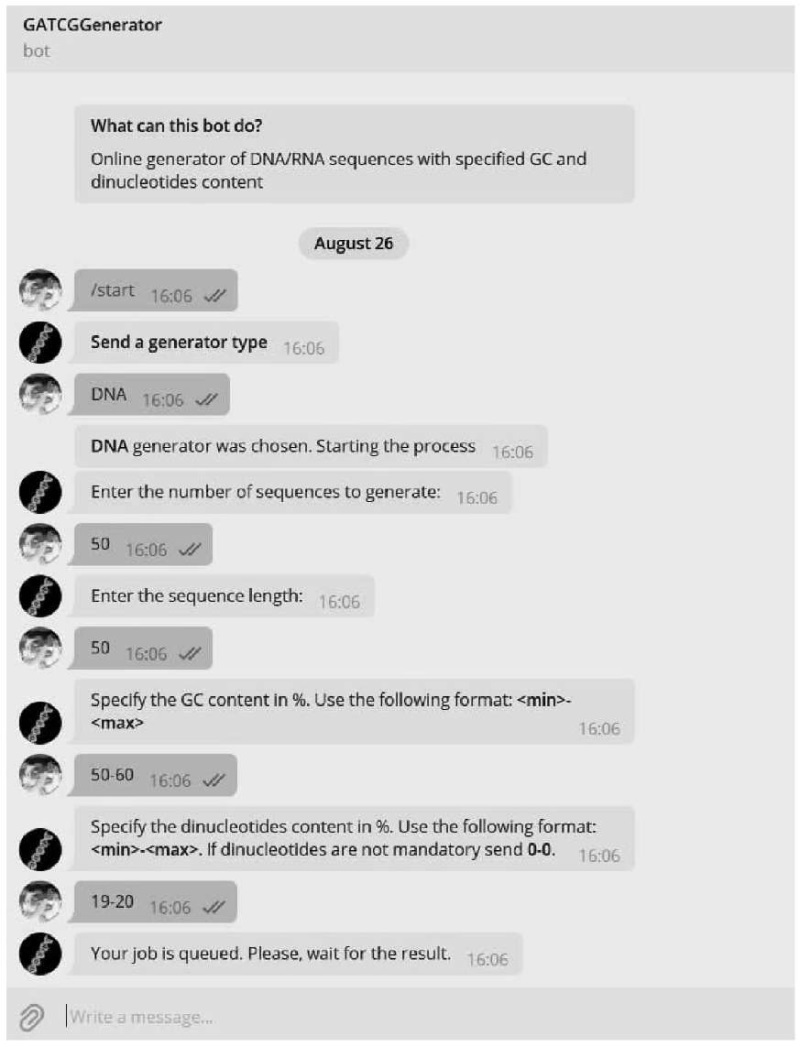
Fig. 1. Example of a user chat in Telegram
In the framework of the presented work, the functionality of random sequence generators and GATCGGenerator were compared. The differences between the input parameters and the specific nucleotide structures obtained as a result of the program were determined (Table 1).
Table 1
Comparison of GATCGGenerator functionality to other nucleotide sequence generators
|
GATCGGenerator [20] |
Nucleotide Sequence Generator8 |
DNA Sequence Tools: Random Sequence Generator9 |
Random DNA Sequence Generator10 |
Random DNA Sequence11 |
Random DNA Generator12 |
|
|
Maximum length (nucleotides) |
5,000 |
1,000,000 |
10,000 |
1,000 |
||
|
Number of sequences |
100 |
1 |
1; 10; 50; 100 |
100 |
||
|
Input GC composition (%) |
+ |
– |
+ |
– |
+ (*) |
|
|
GC composition (%) |
interval |
number |
– |
number |
||
|
Input NN composition (%) |
– |
|||||
|
No homopolymer sites |
+ |
|||||
|
Sequence type |
DNA/RNA |
DNA |
DNA/RNA / Protein |
DNA |
||
|
Output of results |
.CSV file |
Text on the screen |
||||
|
(*) User enters AT composition |
||||||
GATCGGenerator has a broader functionality, it allows the user to specify the number of dinucleotides, create sequences without extended homopolymer sites and repeats that affect the success of the experiment. In existing generators, it is only possible to vary the GC composition.
The program created by the authors of this research generates a given number of quasi-random sequences of nucleotides that do not have homology with natural DNA, but are suitable for molecular biological manipulations.
Research Results. GATCGGenerator allows you to generate specific DNA or RNA sequences from 20 to 5,000 nucleotides long, containing a given number of dinucleotides and not containing homopolymer sites (no more than two identical nucleotides located side by side). More stringent generation conditions can cause a long selection of sequences. As an example, we give a small range of possible content of guanine and cytosine and dinucleotides (e.g., GC composition 45–50 % and NN composition 10–20 %). The operating period of the program for various input data is shown in Table 2.
Table 2
Sequence generation time for different inputs
|
Data inputs |
Time (s) | |||
|
Length |
Number |
GC, % |
NN, % |
|
|
20 |
10 |
50–60 |
20–50 |
3.45 |
|
30 |
10 |
50–60 |
20–50 |
3.91 |
|
20 |
10 |
50–60 |
40–50 |
9.74 |
|
30 |
10 |
50–60 |
40–50 |
9.53 |
|
30 |
10 |
40–50 |
20–20 |
8.80 |
|
1,000 |
100 |
45–50 |
40–50 |
11.49 |
|
2,000 |
100 |
45–50 |
10–20 |
240.25 |
|
5,000 |
100 |
50–60 |
20–50 |
11.57 |
GATCGGenerator, through more stringent sequence generation conditions, removes the limitations of known DNA generators and creates quasi-random sequences of nucleotides depending on the indicated input parameters. You can specify the required number of sequences, their length, GC composition and dinucleotide content, as well as the nature of the nucleic acid (DNA or RNA). Specifically, sequences created by GATCGGenerator can be used in DNA steganography, applied to protect and transmit information through hiding the message content in the nucleotide
sequence [3].
The proposed software solution (GATCGGenerator) provides obtaining a set of quasi-random sequences of nucleotides depending on user-defined input parameters (type of nucleic acid, sequence length, GC and dinucleotide composition). GATCGGenerator excludes the presence of any nucleotide repeats and homopolymer sites longer than three elements. The generated sequences can be used as service or masking sequences (e.g., in DNA steganography) and are suitable for any non-biological enzymatic manipulations. It is possible to generate numerous artificial nucleotide sequences and use them to create a universal oligotheca suitable for multiple encoding of non-biological data and their long-term storage.
The data presented in Table 3 summarizes the results of the program. For a certain type of nucleic acid (in this case, DNA), the following data is shown: the content of dinucleotides (NN %), the number of generated sequences, their length (nucleotides — nt), and GC composition.
Table 3
Examples of short sequences differing in length, GC composition, and dinucleotide content (%)
|
Input parameters |
Nucleotide sequence, 5’→3’ |
Output parameters |
||||||
|
Type |
Number |
Length, nt |
GC, % |
NN, %* |
Length, nt |
GC, % |
NN, %* |
|
|
DNA |
5 |
30 |
41–50 |
20 |
CTGG**TATATCGGAATCATATCGCGCAGTGT |
30 |
46.7 |
20.0 |
|
AATCAGCTAGTAGGACGCAGTAGTGAATCA |
30 |
43.3 |
20.0 |
|||||
|
GAATGTAGTCCTAGGCACATACTACGTAGC |
30 |
46.7 |
20.0 |
|||||
|
AGTTGCACTGAAGTCTATGATCTGGCATGC |
30 |
46.7 |
20.0 |
|||||
|
GACACACTACTATGGACGTGAGGCACTTAC |
30 |
50.0 |
20.0 |
|||||
|
5 |
51–60 |
TCAGCTCAGCGCCAATCGAGCTTATAGTGC |
30 |
53.3 |
20.0 |
|||
|
GAGGCTATCGTCAAGCATAGACCGTGTGCT |
30 |
53.3 |
20.0 |
|||||
|
GACTCAGTAGCTGCTCCGGACATACAGCCT |
30 |
56.7 |
20.0 |
|||||
|
TCGCGCGTTAGACTTAGGTCTCATCGCAGC |
30 |
56.7 |
20.0 |
|||||
|
ACGCTCACAGGAGTTCGCATCGAACGATGC |
30 |
56.7 |
20.0 |
|||||
|
5 |
41–50 |
0 |
ACGACAGTGATATAGCACGACGTGCTCATA |
30 |
46.7 |
0.0 |
||
|
GACTACATCTGATAGTACACGTGCTGCACT |
30 |
46.7 |
0.0 |
|||||
|
TCTATCTCTGCTAGAGCGCTCGTCACTCTA |
30 |
50.0 |
0.0 |
|||||
|
TCTGATCTACTATAGCGATACGTGAGAGTG |
30 |
43.3 |
0.0 |
|||||
|
ACACATATATCGACGCACGCGTCGTAGTAC |
30 |
50.0 |
0.0 |
|||||
|
5 |
50 |
41–60 |
20 |
TGCATGACCATGCTTGCGGTAGACATTCAGACGCGCGAATAGTAGGACGA |
50 |
52.0 |
20.0 |
|
|
GCATACGAGTGGCATACATATTAGACTATACGGTAGTGCATATGGTGCAA |
50 |
42.0 |
20.0 |
|||||
|
CTGAGACTCCTCTCTGTGGAGCTCCTAGTACCGTCACGCGTGCTCTGAAG |
50 |
58.0 |
20.0 |
|||||
|
CTGTGTGAACATACGATGCATTCTCATCTCGGTATGGCTGAAGTGCACAT |
50 |
46.0 |
20.0 |
|||||
|
GCGCTGACGTCATGGTTCATACCAATGTAGCATGATGTGCGATAGGCACA |
50 |
50.0 |
20.0 |
|||||
|
* NN shows the fraction (%) of the dinucleotides contained in the nucleotide sequence. ** Dinucleotides are highlighted in bold. |
||||||||
The obtained 30-nucleotide sequences were tested using the Blast tool from NCBI. The absence of 100% homology with known DNA sequences of living organisms was identified. The maximum coincidence was observed for the generated sequences with a length of 25 nucleotides (similarity is about 80 %). This indicates the ability of the GATCGGenerator to generate non-biological nucleotide sequences with high efficiency. It can be assumed that the sequences generated in this way do not have an absolute coincidence with the nucleotide fragments of living organisms.
In this case, special DNA-oligonucleotides of artificial origin containing informative and service parts can be used as a convenient information carrier. Recently, the authors of this work have proposed the use of NYRN-oligonucleotides [14] consisting of:
- internal part (YR)n encoding the encrypted information;
- service (auxiliary) parts S1 and S2 flanking sequence (YR)n (Fig. 2).

Fig. 2. Structure of NYRN-oligonucleotides:
N — degenerate nucleotides;
Y — pyrimidines (C or T);
R — purines (A or G);
k, n, m — indices corresponding to the length of the part
The length of the sites (n, k and m) may vary, but the structure of the service parts should provide the successful course of amplification reactions (length more than 18 nt, 40–60 % GC composition, absence of homopolymer sites and repeats). GATCGGenerator allows including NN dinucleotides containing identical paired nucleotides (e.g., AA, GG, CC, TT or UU for RNA), which can increase the specificity of molecular hybridization of nucleic acids.
Discussion and Conclusion. Thus, based on the results of the scientific investigation performed, a software solution (GATCGGenerator) has been proposed, which, in comparison to traditional approaches, assumes more stringent conditions for generating sequences. Due to this feature, the limitations of known DNA generators are removed, and quasi-random sequences of nucleotides are formed depending on the specified input parameters. The obtained 30-nucleotide sequences were studied. The test allowed us to establish the absence of 100 % homology with known DNA sequences of living organisms. The generated sequences with a length of 25 nucleotides coincided as much as possible (by about 80 %).
Note also that in order to hide information in NYRN-oligonucleotides, it is required to mix them with masking DNA. Masking sequences should be similar to sequences of NYRN-oligonucleotides, so that when trying to read hidden information, it would be impossible to recognize them without key sequences. The addressee should know the key sequences — primers to the service sites of NYRN-oligonucleotides. The addressee can decipher the transmitted message by isolating informative nucleotide sequences using a polymerase chain reaction followed by sequencing. A set of NYRN- and masking oligonucleotides can be easily obtained using GATCGGenerator, synthesized, and then stored as an oligotheca. To do this, it is enough to determine the optimal NYRN-oligonucleotides with subsequent filling of the oligotheca. In the future, it is planned to conduct laboratory experiments in order to test the proposed method of storing non-biological information and checking the viability of oligotheca obtained using the generator.
1. Nucleotide Sequence Generator. nucleotide-generator.herokuapp.com. URL: https://nucleotide-generator.herokuapp.com/ (accessed: 01.12.2022).
2. DNA Sequence Tools: Random Sequence Generator. molbiotools.com. URL: http://www.molbiotools.com/randomsequencegenerator.html (accessed: 01.12.2022).
3. Random DNA Sequence Generator. faculty.ucr.edu. URL: http://www.faculty.ucr.edu/~mmaduro/random.htm (accessed: 02.12.2022).
4. Random DNA Sequence GenScript. genscript.com. URL: https://www.genscript.com/sms2/random_dna.html (accessed: 04.12.2022).
5. Random DNA Generator. Computer software. URL: http://54.235.254.95/cgi-bin/gd/gdRandDNA.cgi (accessed: 04.12.2022).
6. Anaconda. Anaconda Inc. anaconda.com. URL: https://www.anaconda.com/ (accessed: 20.01.2023).
7. Python telegram bot. github.com. URL: https://github.com/python-telegram-bot/python-telegram-bot (accessed: 01.12.2022).
References
1. Malinetski GG, Mitin NA, Naumenko SA. Nanobiology and Synergetics. Problems and Ideas. Part 2. Keldysh Institute Preprints. 2005;29:1–26. URL: http://mi.mathnet.ru/ipmp722 (accessed: 01.06.2023).
2. Katz E. (ed) DNA- and RNA-Based Computing Systems, 1st ed. Weinheim: Wiley-VCH; 2021. 408 p.
3. Ceze L, Nivala J, Strauss K. Molecular Digital Data Storage Using DNA. Nature Reviews Genetics. 2019;20:456– 466. https://doi.org/10.1038/s41576-019-0125-3
4. Kaundal AK, Verma AK. DNA Based Cryptography: A Review. International Journal of Information and Computation Technology. 2014;4(7):693–698.
5. Aquilanti L, Clementi F, Landolfo S, Nanni T, Palpacelli S, Tazioli A. A DNA Tracer Used in Column Tests for Hydrogeology Applications. Environmental Earth Sciences. 2013;70:3143–3154. https://doi.org/10.1007/s12665-013-2379-y
6. Zhirnov V, Zadegan RM, Sandhu GS, Church GM, Hughes W. Nucleic Acid Memory. Nature Materials. 2016;15:366–370. https://doi.org/10.1038/nmat4594
7. Yetisen AK, Davis J, Coskun AF, Church GM, Seok Hyun Yun. Bioart. Trends in Biotechnology. 2015;33(12):724– 734. https://doi.org/10.1016/j.tibtech.2015.09.011
8. Na D. DNA Steganography: Hiding Undetectable Secret Messages within the Single Nucleotide Polymorphisms of a Genome and Detecting Mutation-Induced Errors. Microbial Cell Factories. 2020;19(128):1–9. https://doi.org/10.1186/s12934-020-01387-0
9. Shuhong Jiao, Goutte R. Code for Encryption Hiding Data into Genomic DNA of Living Organisms. In: Proc. 9th International Conference on Signal Processing. Beijing: IEEE; 2008. P. 2166−2169. https://doi.org/10.1109/ICOSP.2008.4697576
10. Masanori Arita. Writing Information into DNA. In book: N. Jonoska, G. Păun, G. Rozenberg (eds). Aspects of Molecular Computing. Lecture Notes in Computer Science. Berlin, Heidelberg: Springer; 2004. P. 23–35. https://doi.org/10.1007/978-3-540-24635-0_2
11. Church GM, Yuan Gao, Sriram Kosuri. Next-Generation Digital Information Storage in DNA. Science. 2012;337(6102):1628. https://doi.org/10.1126/science.1226355
12. KA Schouhamer Immink, Kui Cai. Design of Capacity-Approaching Constrained Codes for DNA Based Storage Systems. IEEE Communications Letters. 2018;22(2):224–227. https://doi.org/10.1109/LCOMM.2017.2775608
13. Nozomu Yachie, Kazuhide Sekiyama, Junichi Sugahara, Yoshiaki Ohashi, Masaru Tomita. Alignment-Based Approach for Durable Data Storage into Living Organisms. Biotechnology Progress. 2007;23(2):501–505. https://doi.org/10.1021/bp060261y
14. Garafutdinov RR, Sakhabutdinova AR, Slominsky PA, Aminev FG, Chemeris AV. A New Digital Approach to SNP Encoding for DNA Identification. Forensic Science International. 2020;317:110520. https://doi.org/10.1016/j.forsciint.2020.110520
15. Ailenberg M, Rotstein OD. An Improved Huffman Coding Method for Archiving Text, Images, and Music Characters in DNA. BioTechniques. 2009;47(3):747–754. https://doi.org/10.2144/000113218
16. Doricchi A, Platnich CM, Gimpel A, Horn F, Earle M, Lanzavecchia G, et al. Emerging Approaches to DNA Data Storage: Challenges and Prospects. ACS Nano. 2022;16(11):17552–17571. https://doi.org/10.1021/acsnano.2c06748
17. Sakhabutdinova AR, Mikhailenko KI, Garafutdinov RR, Kiryanova OYu, Sagitova MA, Sagitov AM, et al. Non-Biological Application of DNA Molecules. Biomics. 2019;11(3):344–377. https://doi.org/10.31301/2221-6197.bmcs.2019-28
18. Garafutdinov RR, Chemeris DA, Sakhabutdinova AR, Chemeris AV, Kiryanova OYu, Mikhaylenko CI. Encoding of Non-Biological Information for its Long-Term Storage in DNA. Biosystems. 2022;(215–216):104664. https://doi.org/10.1016/j.biosystems.2022.104664.9
19. Kiryanova OYu, Kiryanova II, Garafutdinov RR, Chemeris DA, Gubaidullin IM. GATCGGenerator. Certificate of Software Registration No. RU 2021667097. 2021. (In Russ.)
20. Borzov EA, Marakhonov AV, Ivanov MV, Drozdova PB, Baranova AV, Skoblov MYu. RANDTRAN: Random Transcriptome Sequence Generator that Accounts for Partition Specific Features in Eukaryotic mRNA Datasets. Molecular Biology. 2014;48:749–756. https://doi.org/10.1134/S0026893314050021
21. Harris CR, Millman KJ, van der Walt SJ, Gommers R, Virtanen P, Cournapeau D, et al. Array Programming with NumPy. Nature. 2020;585:357–362. https://doi.org/10.1038/s41586-020-2649-2
About the Authors
O. Yu. KiryanovaRussian Federation
Olga Yu. Kiryanova, Teaching assistant of the Department of Digital Technologies and Modeling
1, Kosmonavtov St., Ufa, 450064
R. R. Garafutdinov
Russian Federation
Ravil R. Garafutdinov, Cand.Sci. (Chemistry), Head of the Laboratory of Physico-Chemical Methods of Analysis of Biopolymer
71, Oktyabrya Av., Ufa, 450054, Bashkortostan Rep.
I. M. Gubaydullin
Russian Federation
Irek M. Gubaydullin, Dr.Sci. (Phys.-Math.), Professor, Head of the Laboratory of Mathematical Chemistry
141, Oktyabrya Av., Ufa, 450075, Bashkortostan Rep.
A. V. Chemeris
Russian Federation
Aleksei V. Chemeris, Dr.Sci. (Biology), Professor, Chief Research Fellow
71, Oktyabrya Av., Ufa, 450054, Bashkortostan Rep.
Review
For citations:
Kiryanova O.Yu., Garafutdinov R.R., Gubaydullin I.M., Chemeris A.V. GATCGGenerator: New Software for Generation of Quasirandom Nucleotide Sequences. Advanced Engineering Research (Rostov-on-Don). 2023;23(3):296-306. https://doi.org/10.23947/2687-1653-2023-23-3-296-306
JATS XML














































