Scroll to:
Computer Program for Primer Design for Loop-Mediated Isothermal Amplification (LAMP)
https://doi.org/10.23947/2687-1653-2024-24-1-98-108
EDN: TRWNOM
Abstract
Introduction. To date, numerous methods of nucleic acid amplification have been proposed, and each method has a number of advantages and disadvantages. One of the most popular methods is Loop-mediated isothermal AMPlification (LAMP). Unlike thermocyclic reactions, such as PCR (polymerase chain reaction), which require three temperature changes and expensive equipment, in LAMP, the entire reaction takes place at one and the same temperature and at the maximum rate possible. An important component of LAMP-amplification is primers (usually 20–25 nucleotides), which need to be matched to a specific part of the nucleotide sequence. It is known that DNA sequence contains four nucleotides: A — adenine and T — thymine, G — guanine and C — cytosine. There is a huge variety of permutations of these nucleotides, and it is practically impossible to analyze such a large amount of data manually. Therefore, there is a need to use modern computer technologies. More than 150 computer programs have been proposed for the design of PCR primers, while for LAMP-primers there are less than 10 of them, and each of them has a number of drawbacks, e.g., in terms of the length of the analyzed site. Therefore, this work is aimed at developing a new domestic computer program for the design of specific primers for LAMP.
Materials and Methods. The primer search algorithm was based on a linear search for a substring in a string, taking into account the criteria of primer selection for LAMP. The program complex of LAMP-primer design was implemented in Python programming language. The bioPython library was used to work with various DNA and RNA, and the Qt framework was used to develop the interface.
Results. A modification of the direct sampling method using a stencil approach was proposed, taking into account the GC composition and annealing temperature of primers depending on their structure. A software package with a friendly interface was developed. It took into account the design criteria of primers: certificates of registration of computer programs (LAMPrimers iQ No. 2022617417 dated April 20, 2022, LAMPrimers iQ_loop No. 2023662840 dated June 14, 2023) were received. The program is in the public domain at https://github.com/Restily/LAMPrimers-iQ
Discussion and Conclusion. The developed software packages can be used for research and analysis in molecular biology and genetics, to create diagnostic test systems that provide high sensitivity and reliability of detection of specific DNA and RNA. The software packages can be used in research institutes and laboratories engaged in the amplification of nucleic acids. The results of evaluating the selected sets of primers for the LAMP reaction were tested, and the effectiveness of working sets using the LAMPrimers iQ program was experimentally proven by the example of the detection of genetic material of the SARS-CoV-2 coronavirus.
For citations:
Akhmetzianov L.U. Computer Program for Primer Design for Loop-Mediated Isothermal Amplification (LAMP). Advanced Engineering Research (Rostov-on-Don). 2024;24(1):98-108. https://doi.org/10.23947/2687-1653-2024-24-1-98-108. EDN: TRWNOM
Introduction. Nucleic acid amplification is a valuable molecular tool not only in fundamental research, but also in applied areas, such as the diagnosis of infectious diseases, hereditary pathologies, establishing kinship, etc. Currently, amplification methods are intensively developing, and their application areas are expanding. The most popular and most frequently used amplification method is polymerase chain reaction (PCR) [1]. PCR is a reaction that takes place under three different temperature conditions: denaturation (95°C), annealing of primers (from 50° to 60°C), elongation (72°C). To quickly change these modes, a special device is needed — a DNA thermocycler [2]. At this, temperature variation in the amplifier does not occur instantly, but starts only when the desired temperature is reached, and this causes artificial containment of the reaction. As a rule, the duration of PCR is 1–1.5 hours.
The second most popular amplification method is loop-mediated isothermal amplification (LAMP) [3]. A water bath or thermostat is sufficient for LAMP, since the reaction takes place at one and the same temperature, and the first results can be seen in 15 minutes.
For both LAMP and any other type of amplification, the key component is primers, which are short sequences of nucleic acid. They serve as a starting point for increasing copies of a specific region of DNA. It is the primers that determine which DNA sequence will be copied.
The main difference of LAMP implementation is the number of primers. For a conventional LAMP, at least four primers are required (two external, two internal), while for a conventional PCR, two are sufficient (direct, reverse).
To increase the specificity and accuracy of the reaction, it is important to select the right primers. For the automatic selection of primers for PCR, more than 150 different computer programs have been developed that provide the selection of primers for any modifications of this reaction [4]. However, there are very few such programs for LAMP, no more than ten, and only two of them are available online. These programs also have a number of disadvantages, such as restrictions on the length of the analyzed sequence; they do not exclude the possibility of formation of homo- and heterodimers of primers, repeats of nucleotides in one primer. And none of the programs take into account the close arrangement of primers in one set, which, in turn, reduces the quality of primers and the accuracy of reaction results [5].
Thus, an urgent task is to develop a new computer program to select (model) high-quality sets of primers for LAMP with tougher conditions for choosing primers for nucleotide sequences of any length.
Materials and Methods. The authors of [3] proposed using two external, F3 (Forward), B3 (Backward), and two internal primers, FIP (Forward Inner Primer), BIP (Backward Inner Primer). It was assumed that the internal primers had double length (FIP: F1c/F2, BIP: B1c/B2) and were annealed at four regions of the nucleotide sequence. Schematically, the location of LAMP primers can be seen in Figure 1.
External primers are only needed at the initial stage. They are designed to limit the analyzed region of the nucleotide sequence and form a single-stranded structure of this region. A pair of internal primers, F1c and B1c, start their work already at the second stage, as they are annealed after the formation of new DNA chains.
Later, the same authors proposed a modified method offering the use of not four, but six primers, annealed already at eight regions of the target nucleotide sequence [7]. It was proposed to add two more loop primers (Loop B, Loop F), which should react at the third stage after the formation of a dumbbell-like DNA structure and anneal between regions F1/F2 and B1/B2, respectively. The use of additional primers implies an increase in sensitivity and reliability of the reaction.
Any amplification reaction has its own sensitivity threshold, and the spread of this indicator is very large due to the fact that numerous factors affect the course of both PCR and LAMP. Some papers have noted that LAMP is significantly more sensitive than PCR. The authors of [8], e.g., claim that LAMP is 10 times more sensitive than PCR. The authors of other studies have found that LAMP is 100 times more sensitive than PCR [9], and in some investigations, this indicator reaches 1,000 times [10].
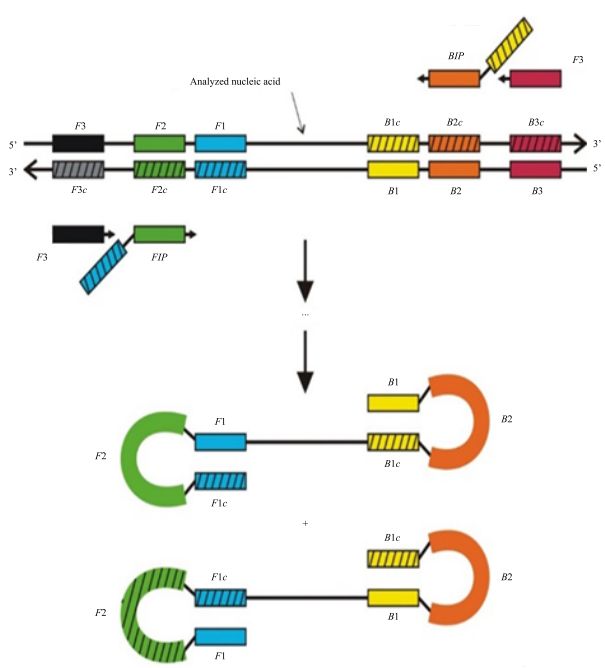
Fig. 1. Schematic arrangement of the annealing regions of external and internal primers for LAMP [6]
In addition to sensitivity, any amplification reaction has another equally important indicator — its specificity. And here, questions have recently begun to arise about the LAMP method [11], including due to the emergence of the so-called primer homo- and heterodimers, which are more difficult to exclude in this reaction than in PCR, because of the larger number of primers used and their increased length [12].
For successful amplification, it is necessary to select the right primers. When using the LAMP method, the main difficulty is in modeling primers with account for all recommended conditions, namely:
- length of the primer (18–35 nucleotides for external primers, 30–55 nucleotides for internal ones);
- content of guanine (G) and cytosine (C) (GC composition ranging from 40 to 60%);
- optimal primer annealing temperature (55–65°C);
- close arrangement of primers in one set: the average size of the amplicon (120–220 bp);
- exclusion of the formation of dimers of primers;
- elimination of nucleotide repeats in one primer (no more than three).
Table 1 provides brief characteristics and features of the most popular LAMP primer design programs [5].
Table 1
Brief characteristics of popular computer programs for LAMP primer design
|
|
Development language |
Length of the analyzed region, nucleotides |
Selection of loop primers |
Configuration of loop primers |
Graphical interface |
Free access |
|
Primer Explorer (V4, V5), Japanese company Eiken Chemical Co. LTD, Tokyo |
Java |
up to 2,000 |
+ |
no |
yes |
yes |
|
FastPCR, Finnish company Primer Digital Ltd |
Java |
from 12 up to 500 |
– |
no |
no |
no |
|
С, CUDA С, Perl OS Linux |
unlimited |
+ |
+ |
no |
yes |
|
|
|
up to 15,000,000 |
+ |
only Tm |
yes |
no |
|
|
Python |
|
– |
– |
– |
no access |
|
|
Java |
from 100 up to 2,000 |
+ |
– |
yes |
yes |
|
|
Python 3.10 |
unlimited |
+ |
yes |
yes |
yes |
The design of primers for LAMP is a very difficult task and requires the development of a special computer program with proper functionality, taking into account all recommended conditions, and with the possibility of an extended selection of primers and a user-friendly interface.
The computer program for primer design is developed in the Python programming language. This language has the bioPython library, which allows working with nucleotide sequences, as well as the Qt framework for interface development.
Research Results. With account for the structural features of nucleotide sequences and the criteria for selecting LAMP primers, a modification of the direct sampling method using a stencil approach has been proposed. It considers the GC composition, the annealing temperature of the primers, and reducing the complexity of the sampling.
As is known, the GC composition of primers should be in the range from 40 to 60%. This is one of the important criteria for selecting LAMP primers, which depends on the sequence being analyzed, the length of the primer, and it partially affects the annealing temperature (Тm, °С).
In this paper, the following formula is used to calculate the annealing temperature of primers:
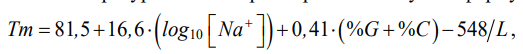 (1)
(1)
where [Na+] — molar concentration of sodium ions; (%G + %C) — GC composition in the analyzed sequence, expressed as a percentage; L — length of the primer. It was based on a well-known dependence:
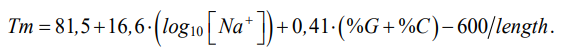 (2)1
(2)1
Formula (1) was selected empirically, the calculated values were compared to the values obtained through the convenient OligoAnalyzer2 utility, which provided a high-quality selection of primers for all types of nucleotide acid amplification. Through determining the length of the primers, GC composition and annealing temperature, all possible primers can be found in a nucleotide sequence of any length that will meet the specified criteria.
If we imagine the primer as a substring, and the analyzed nucleotide sequence as a longer string, this task can be represented as a search of all possible options (direct search), but complicating it through calculating the GC composition and annealing temperature of the primers.
Figure 2 shows a complete block diagram of the direct search algorithm, taking into account:
- length of the primers (bp);
- GC-composition, %;
- annealing temperature of the primers, Tm, °C;
- homodimers on both DNA strands.
The total complexity of the modified algorithm in the worst case is O(m×n), where n — length of the primer, m — length of the nucleotide sequence. It should be understood that the running time of the algorithm directly depends on how often the nucleotide fragments that meet the requirements are found.
Table 2 shows the search data for all possible primers in nucleotide sequences of different structures over time.
Further, it is necessary to form sets from all the primers found, taking into account the close distance between the primers, the heterodimeric properties in one set, as well as the minimum temperature difference between the annealing of the primers.
The design scheme for forming primers into the LAMP sets is shown in Figure 3.
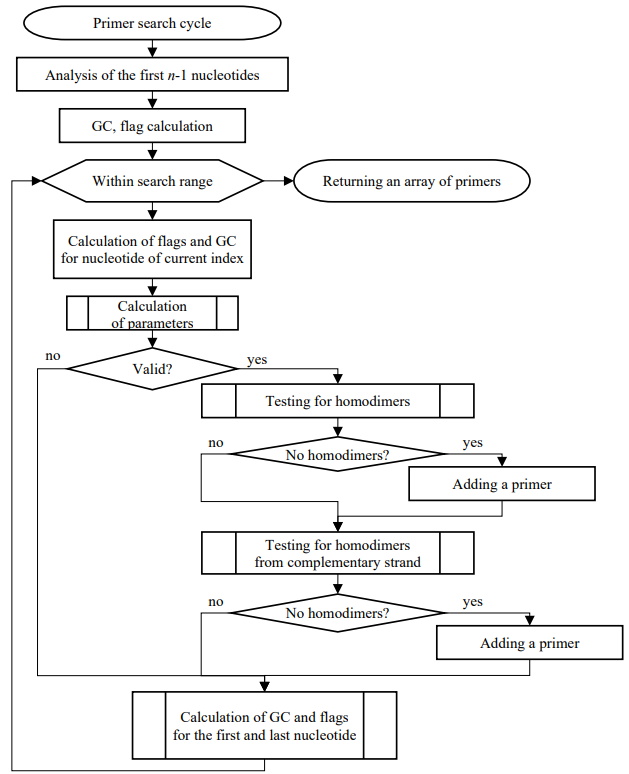
Fig. 2. Complete flowchart of the algorithm
Table 2
Time spent searching for valid primers in nucleotide sequences of different lengths
|
Genome name |
Nucleotide sequence size, bp |
Search for primers, s |
|
SARS-CoV-2 |
29,903 |
0.31 |
|
Escherichia virus T4 |
168,903 |
1.73 |
|
Mycoplasma |
580,076 |
5.43 |
|
Helicobacter pylori |
1,624,458 |
18.11 |
|
Escherichia coli |
4,641,652 |
71.68 (1.2 min) |
|
Caenorhabditis |
100,286,401 |
1,082.53 (18 min) |
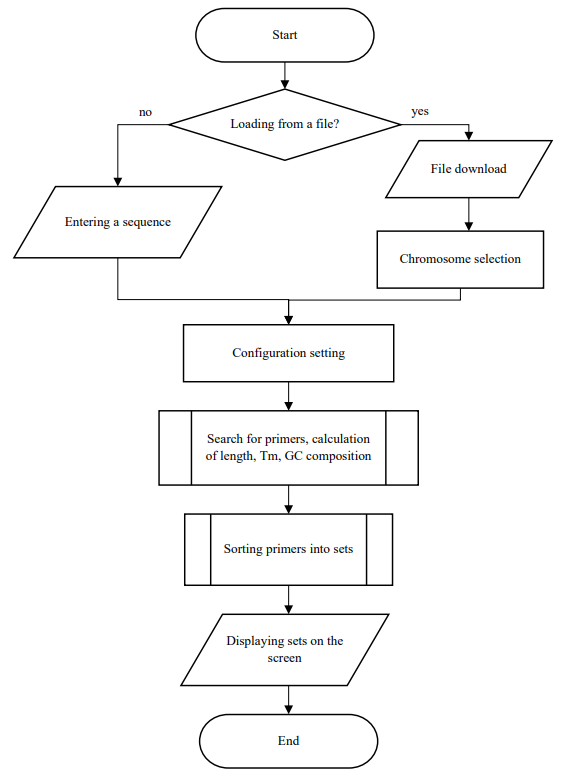
Fig. 3. Block diagram of the formation of primer sets for LAMP
At the input, the program reads the nucleotide sequence, either the desired file is loaded, or a fragment is inserted through the clipboard. Next, configurations are set, such as primer length, GC composition, primer annealing temperature, and temperature difference in one set. Then, all possible parameters satisfying the set configurations are searched, the selected primers are sorted into sets and displayed to the user.
The operation of the computer program:
- Uploading a file (simple text format, FASTA format, GenBank), or a fragment of a sequence via the clipboard.
- Search for all possible primers: primers are combined according to the following criteria:
- length of the analyzed region;
- distances between primers (F3/F2 — 1–10 nucleotides, F2/F1c — 10–25 nucleotides, F1c/B1c — 0-30 nucleotides);
- temperature difference of primer annealing (<3);
- heterodimers.
If all of the above conditions are met, the set is considered to be working.
- Displaying simulated primer sets to the user's screen and/or saving to a file.
The LAMP program is registered in the Register of Computer Programs under the name LAMPrimers iQ, No. 2022617417 on April 20, 2022, and LAMPrimers iQ-loop, No. 2023662840 on June 14, 2023. The program code is publicly available3.
The developed software product has a user-friendly and intuitive interface, which can be used directly by end users — experimenters engaged in LAMP.
Discussion and Conclusion. The number of primer sets issued depends on the specified search parameters. The stricter the parameters, the fewer sets will be found. In case of hard restrictions, the program may not output a single set. The number of primer sets with different selection parameters for the genome of the bacteriophage lambda [13], whose length is ~48,500 nucleotides, is shown in Table 3.
For relatively short nucleotide sequences (up to 2,000 nucleotides), the selection of primers takes less than a second. With increasing sequence length, the duration of the primer search grows exponentially.
Table 3
Number of the primer sets for the bacteriophage lambda genome, depending on the set selection parameters
|
GC, % |
ΔTm, °C |
Maximum length of amplified region, bp |
||
|
300 |
230 |
160 |
||
|
40–60 |
5 |
213 |
119 |
3 |
|
2 |
198 |
184 |
3 |
|
|
45–55 |
5 |
132 |
80 |
0 |
|
2 |
116 |
64 |
0 |
|
|
50–60 |
5 |
195 |
185 |
4 |
|
2 |
181 |
164 |
4 |
|
|
55–65 |
5 |
160 |
147 |
4 |
|
2 |
134 |
105 |
0 |
|
Figure 4 shows the effect of the length of the nucleotide sequence on the duration of the selection of primer sets. The data for the nucleotide sequence of the bacteriophage lambda on a laptop with the parameters are as follows: Intel(R) Core(TM) i7-10750H CPU, 2.60GHz, 6 cores. 16 GB RAM. The parameters of the soft selection of primers are indicated (40–60 % GC, ΔTm = 5, length of the analyzed region is up to 300 bp).
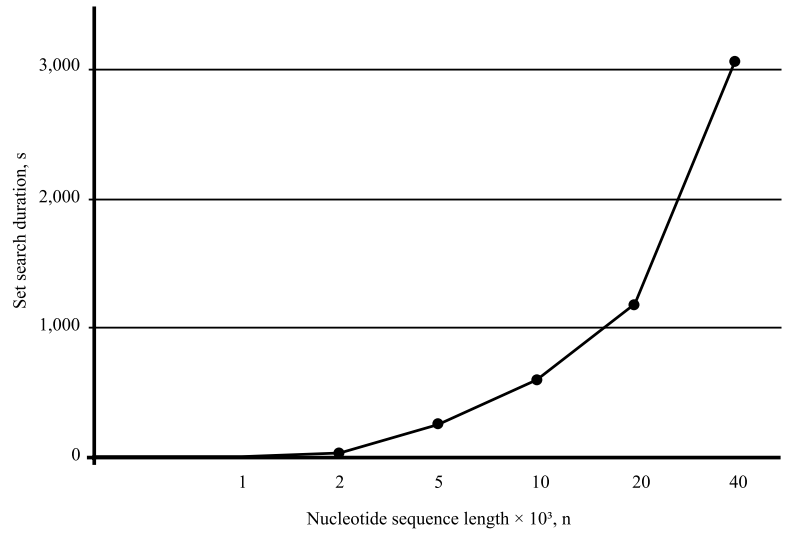
Fig. 4. Effect of length of nucleotide sequence on duration of selection of primer sets
It should be noted that the duration of the search for primers depends on the power of the computer.
To compare and determine the quality of the simulated primer sets, a number of field experiments were conducted to detect the RNA of the SARS-CoV-2 coronavirus, whose length was ~ 30,000 nucleotides. To do this, sets of LAMP primers were selected for the same region of the nucleotide sequence of the coronavirus using the LAMPrimers iQ program and two popular and accessible online utilities from New England Biolabs (NEB LAMP)4 and PrimerExplorer5. The designations L, N and P correspond to the sets of primers LAMPrimers iQ, NEB LAMP Primer Design and PrimerExplorer; “+” — samples contained RNA of SARS-CoV-2 coronavirus, “–” — samples did not contain nucleic acids. Figure 5 shows the curves of this experiment.
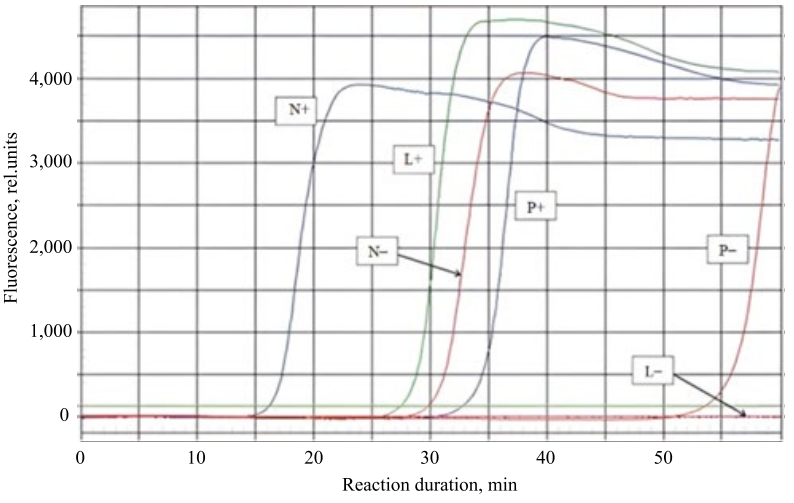
Fig. 5. Graph of the comparative experiment (the author’s figure):
L — primers obtained using LAMPrimers Iq, N — NEB LAMP Primer Design,
P — PrimerExplorer, samples contained RNA of SARS-CoV-2 coronavirus,
“–” — control samples without matrix (did not contain nucleic acids)
The primers obtained through PrimerExplorer, showed the latest rise in amplification curves (P+) compared to NEB LAMP (N+) and LAMPrimers iQ (L+). The primers obtained through LAMPrimers iQ, provided a later rise in the amplification curves (L+) compared to (N+). However, samples that did not contain virus RNA (P–), showed a later rise compared to the set (N–), while (L–) showed no rises even after 50 minutes, thereby providing the highest reliability of viral RNA detection.
The performed experiments showed a higher accuracy and specificity of the primer sets selected through LAMPrimers iQ computer program, caused by a decrease in the reaction rate with negative control samples. The amplification curves had later rises, or did not have them at all, even after 50 minutes of reaction time.
1. Oligo Calc: Oligonucleotide Properties Calculator. URL: http://biotools.nubic.northwestern.edu/OligoCalc.html (accessed: 10.12.2023).
2. OligoAnalyzer™ Tool. URL: https://eu.idtdna.com/pages/tools/oligoanalyzer?returnurl=%2Fcalc%2Fanalyzer (accessed: 10.12.2023).
3. LAMPrimers-IQ. URL: https://github.com/Restily/LAMPrimers-iQ/blob/main/lamp/start_lamp.py (accessed: 10.12.2023).
4. NEB LAMP Primer Design Tool. URL: https://lamp.neb.com/#!/ (accessed: 24.11.2023).
5. LAMP primer designing software Primer Explorer. URL: http://primerexplorer.jp/e (accessed: 25.11.2023).
References
1. Gilmiyarova FN, Kolotyeva NA, Gusyakova OA, Sidorova IF. Polymerase Chain Reaction. History of Discovery. New Stage of Development. Konsilium. Laboratornaya diagnostika. 2017;154(4):17–21. URL: https://cyberleninka.ru/article/n/polimeraznaya-tsepnaya-reaktsiya-istoriya-otkrytiya-novyy-etap-razvitiya/viewer (accessed: 05.11.2023). (In Russ.).
2. Garafutdinov RR, Baymiev AKh, Maleev GV, Alexeyev YaI, Zubov VV, Chemeris DA, et al. Diversity of PCR Primers and Principles of Their Design. Biomics. 2019;11(1):23–70. URL: https://biomicsj.ru/upload/iblock/ddf/bmcs19111023_1_.pdf?ysclid=lruepfy4zf297918053 (accessed: 05.11.2023).
3. Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, et al. Loop-Mediated Isothermal Amplification of DNA. Nucleic Acids Research. 2000;28(12):e63 https://doi.org/10.1093/nar/28.12.e63
4. Chemeris DA, Kiryanova OYu, Gubaydullin IM, Chemeris AV. Design of Primers for Polymerase Chain Reaction (Brief Review of Software and Databases). Biomics. 2016;8(3):215–238. URL: https://biomicsj.ru/upload/iblock/b05/3.pdf (accessed: 05.11.2023).
5. Akhmetzianova LU, Davletkulov TM, Garafutdinov RR, Gubaydullin IM. Application of the Aho-Korasik Algorithm for the Selection of Primers for Loop Isothermal Amplification. Mathematical Biology and Bioinformatics. 2022;17(2):250–265. https://doi.org/10.17537/2022.17.250
6. Akhmetzianova LU, Davletkulov TM, Sakhabutdinova AR, Chemeris AV, Gubaydullin IM, Garafutdinov RR. LAMPrimers iQ: New Primer Design Software for Loop-Mediated Isothermal Amplification (LAMP). Analytical Biochemistry. 2023;684:115376. https://doi.org/10.1016/j.ab.2023.115376
7. Nagamine K, Hase T, Notomi T. Accelerated Reaction by Loop-Mediated Isothermal Amplification Using Loop Primers. Molecular and Cellular Probes. 2002;16(3):223–229. https://doi.org/10.1006/mcpr.2002.0415
8. Zhiyong Chen, Yuxue Liao, Xuemei Ke, Jie Zhou, Yixiong Chen, Lulu Gao, et al. Comparison of Reverse Transcription Loop-Mediated Isothermal Amplification, Conventional PCR and Real-Time PCR Assays for Japanese Encephalitis Virus. Molecular Biology Reports. 2011;38(6):4063–4070. https://doi.org/10.1007/s11033-010-0525-0
9. Nkere CK, Oyekanmi JO, Silva G, Bömer M, Atiri GI, Onyeka J, et al. Chromogenic Detection of Yam Mosaic Virus by Closed-Tube Reverse Transcription Loop-Mediated Isothermal Amplification (CT-RT-LAMP). Archives of Virology. 2018;163(4):1057–1061. https://doi.org/10.1007/s00705-018-3706-0
10. Wang C, Shen X, Lu J, Zhang L. Development of a Reverse Transcription-Loop-Mediated Isothermal Amplification (RT-LAMP) System for Rapid Detection of HDV Genotype 1. Letters in Applied Microbiology. 2013;56(3):229–235. https://doi.org/10.1111/lam.12039
11. Hardinge P, Murray JAH. Lack of Specificity Associated with Using Molecular Beacons in Loop Mediated Amplification Assays. BMC Biotechnology. 2019;19(1):55. https://doi.org/10.1186/s12896-019-0549-z
12. Bodulev OL, Sakharov IYu. Isothermal Nucleic Acid Amplification Techniques and Their Use in Bioanalysis. Biochemistry. 2020;85(2):174–196. https://doi.org/10.31857/S0320972520020037
13. Rebolskaya YuA, Vasenko EA, Melnik SV, Sabirov DKh, Sarygina EV. Bacteriophage Lambda as a Model Object in Genetic Research. In: Proc. X Int. Student Electronic Sci. Conf. “Student Scientific Forum-2018”. Saratov: LLC Nauchno-izdatel'skii tsentr “Akademiya Estestvoznaniya”; 2018. URL: https://scienceforum.ru/2018/article/2018002259 (accessed: 09.10.2023). (In Russ.).
About the Author
L. U. AkhmetzianovRussian Federation
Liana U. Akhmetzianova, teaching assistant of the Department of Computer Science and Engineering Cybernetics
1, Kosmonavtov St., Ufa, 450064
Review
For citations:
Akhmetzianov L.U. Computer Program for Primer Design for Loop-Mediated Isothermal Amplification (LAMP). Advanced Engineering Research (Rostov-on-Don). 2024;24(1):98-108. https://doi.org/10.23947/2687-1653-2024-24-1-98-108. EDN: TRWNOM













































